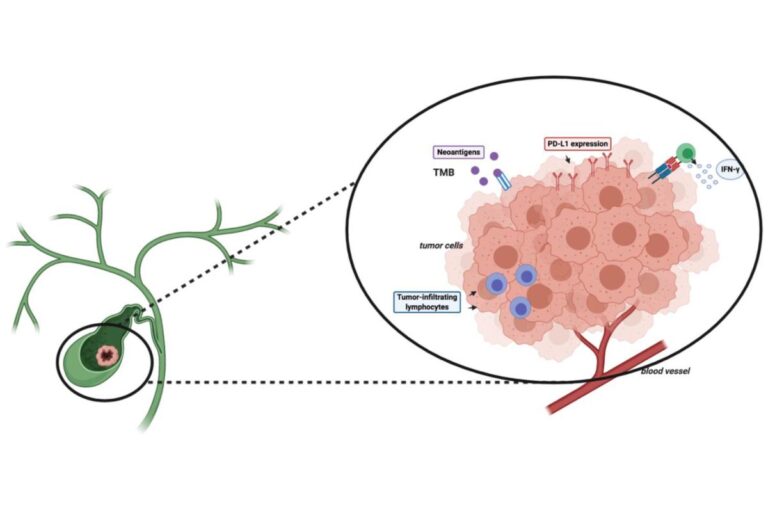
Tumor mutational burden (TMB), microsatellite instability (MSI), and neoantigen prediction have become central molecular readouts in modern immuno-oncology. Each provides a different view of tumor immunogenicity, and together they guide checkpoint inhibitor selection, stratify clinical trial cohorts, and support translational biomarker discovery. As oncology sequencing services continue to expand in sophistication, researchers now have the ability to quantify these biomarkers with far higher precision—directly from whole-exome, whole-genome, or targeted hybrid-capture data.
Below is a detailed overview of the NGS-based strategies used to measure TMB, detect MSI, and predict neoantigens, along with practical considerations for high-confidence interpretation.
Table of Contents
Understanding Tumor Mutational Burden (TMB): Quantifying Tumor Immunogenicity
TMB measures the number of somatic mutations per megabase of coding sequence. Elevated TMB is associated with higher neoantigen load and increased probability of response to immune checkpoint inhibitors.
Modern oncology sequencing services use:
- Whole-exome sequencing (WES) for comprehensive mutation load assessment across >20,000 genes
- Large targeted panels (300–1,500 genes) calibrated to correlate with WES-derived TMB
- Advanced variant-calling pipelines optimized to detect low-frequency somatic mutations in samples with variable tumor purity
Critical technical considerations include minimum sequencing depth, handling FFPE-induced artifacts, and exclusion of germline variants through tumor–normal pairing. Without a paired normal sample, background germline noise can inflate apparent TMB.
In both solid tumors and hematologic malignancies, accurate TMB estimation depends on standardized pipelines—especially when comparing across studies or integrating retrospective datasets.
Microsatellite Instability (MSI): A High-Impact Biomarker for Immunotherapy Response
MSI reflects defects in mismatch repair (MMR) pathways that cause expansions or contractions in microsatellite regions. MSI-high tumors exhibit extensive genomic instability and respond exceptionally well to PD-1 inhibitors.
NGS-based MSI detection offers several advantages over PCR and IHC:
- Genome-wide sensitivity: Thousands of microsatellite loci analyzed simultaneously
- Ability to detect partial or subclonal MSI signatures
- Simultaneous assessment of MMR gene mutations, LOH events, and epigenetic inactivation
- No need for matched normal tissue (depending on algorithm)
Current oncology sequencing services integrate MSI detection directly into somatic variant analysis pipelines. Tools such as MSIsensor, MANTIS, and SigMA evaluate microsatellite length distributions, generating quantitative MSI scores rather than binary classifications.
MSI calling is highly dependent on sequencing quality, read length, and coverage uniformity—making standardized wet-lab and bioinformatic workflows essential.
Neoantigen Prediction: Connecting Mutational Profiles to Immune Recognition
Neoantigens arise when somatic mutations create novel peptides presented on MHC molecules. These peptides can be recognized by T cells, making them key targets in personalized cancer vaccines, adoptive T-cell therapies, and response prediction for immunotherapies.
NGS-based neoantigen prediction pipelines typically follow a multi-step process:
- Somatic variant calling from WES, WGS, or large targeted panels
- HLA typing from tumor or normal DNA
- In silico peptide generation, incorporating missense mutations, frameshifts, or indels
- MHC binding affinity prediction using machine-learning models
- Integration with RNA-seq data to assess expression of neoantigen-generating transcripts
Recent advancements include AI-powered predictors that incorporate structural models of peptide–MHC interactions and TCR engagement probabilities.
Because variant allele frequency, expression levels, and peptide stability all influence immunogenicity, oncology sequencing services increasingly combine WES, RNA-seq, and immune repertoire profiling to improve predictive accuracy.
Best Practices for High-Confidence Immunogenomic Biomarkers
To ensure robust TMB, MSI, and neoantigen insights, researchers should apply:
- Tumor–normal paired sequencing for clean separation of germline and somatic events
- Deep sequencing (>150–250×) for confident low-frequency mutation detection
- Standardized variant-calling pipelines validated against reference datasets
- Integrated multiomic profiling (WES + RNA-seq + HLA typing) for neoantigen prediction
- Rigorous QC on FFPE samples to mitigate artifacts that affect TMB and MSI
These practices are now widely incorporated across advanced oncology sequencing services, enabling consistent biomarker development across diverse tumor types and study designs.
Conclusion
As precision immuno-oncology continues to evolve, TMB, MSI, and neoantigen profiling have become foundational biomarkers. Next-generation sequencing—supported by well-validated wet-lab workflows, multiomic analysis, and machine-learning–driven interpretation—provides the resolution needed to quantify tumor immunogenicity with accuracy. With the growth of comprehensive oncology sequencing services, researchers can now integrate these biomarkers into translational studies, clinical trial design, and therapeutic development with far greater confidence.